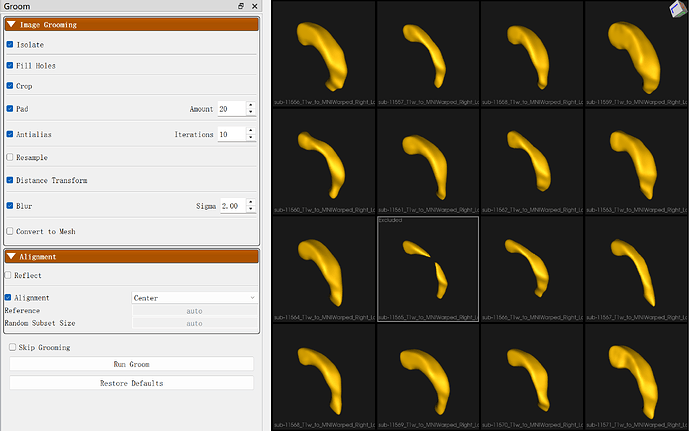
Hello, I encountered such a problem during grooming. The first image is my original ventricle file, and the last one is the file after grooming. You will notice that the ventricle file, which was originally one shape, has become two after grooming. The parameters I used for grooming are shown in the last image. Can this issue be avoided? If not, are there any Python APIs in ShapeWorks that can process the DT to retain only the largest component?
It looks like what is happening here is that the Blur step is blurring enough that the object becomes disconnected. There is an isolate functionality that will isolate the largest connected component and it’s part of Studio’s grooming, but it runs earlier in the pipeline. However, I do not think you want to do this anyway as you will lose half the shape.
Instead what I would recommend if you need this level of smoothing would be to reduce the blurring considerably, enable “Convert to Mesh”, and then use the Mesh Smoothing options, which should be able to smooth the surface without affecting the geometry in this way. I would experiment with both Laplacian and WindowedSinc methods to see which gives the best results for your data.