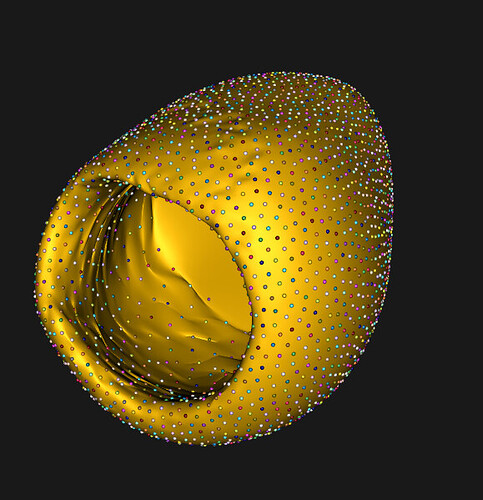
Dear ShapeWorks Support Team,
I am currently using ShapeWorks to build a statistical shape model for a set of 146 right ventricle surface meshes. My goal is to compute a mean shape (template) and then use that mean template to establish consistent topology (correspondence) for all meshes.
The steps I followed are:
I ran an groom(I also did rigid-alignment) and optimization process in ShapeWorks to compute the mean shape of the 146 meshes.
Then, I used this mean template to warp back to each of the original 146 meshes (I believe this step is intended to reconstruct each original mesh by deforming the mean with the deformation field obtained during the correspondence optimization?).
However, I encountered a problem in the second step. When I warp the mean template back to reconstruct the individual meshes, I observe:
Large losses (high reconstruction errors).
The reconstructed meshes appear broken (visually corrupted).
I am unsure why this happens. The optimization seemed to run without errors, and I expected the warping to produce meshes close to the original ones with the same topology.
Could you please help me understand:
Why might the warping result in large errors and broken meshes?
Are there common pitfalls or parameters that I might have set incorrectly?
How can I debug or improve the process?
Thank you for your assistance.
import os
import shapeworks as sw
import numpy as np
# File paths
reference_mesh_path = r"F:\Project_2\3D_DATA\Roberto_SSM\Split_By_Parts\RV\Average_5000_mesh.vtk"
reference_particle_path = r"F:\Project_2\3D_DATA\Roberto_SSM\Split_By_Parts\RV\Average_5000_particles.particles"
output_dir = r"F:\Project_2\3D_DATA\Roberto_SSM\Split_By_Parts\RV\RV_WarpOut"
particles_dir = r"F:/Project_2/3D_DATA/Roberto_SSM/Split_By_Parts/RV/RV_146_particles"
mesh_dir = r"F:/Project_2/3D_DATA/Roberto_SSM/Split_By_Parts/RV/groomed"
# Find matching files
particles_files = [f for f in os.listdir(particles_dir) if f.endswith("_RV_rigid_groomed_local.particles")]
cases = []
for pf in particles_files:
case_prefix = pf.replace("_RV_rigid_groomed_local.particles", "")
mesh_file = f"{case_prefix}_RV_rigid_groomed.vtk"
mesh_path = os.path.join(mesh_dir, mesh_file)
if os.path.exists(mesh_path):
cases.append((os.path.join(particles_dir, pf), mesh_path))
print(f"Found {len(cases)} valid particle-mesh pairs")
# Load reference data
reference_mesh = sw.Mesh(reference_mesh_path)
print(f"Loaded reference mesh: {reference_mesh_path}, Points: {reference_mesh.numPoints()}, Faces: {reference_mesh.numFaces()}")
ref_particles = sw.ParticleSystem([reference_particle_path]).ShapeAsPointSet(0)
print(f"Loaded reference particles: {reference_particle_path}, Particle count: {ref_particles.shape}")
# Create warper object
warper = sw.MeshWarper()
warper.generateWarp(reference_mesh, ref_particles)
# Test warping with reference particles
try:
test_mesh = warper.buildMesh(ref_particles)
if test_mesh.numPoints() == 0 or test_mesh.numFaces() == 0:
print("Test buildMesh failed after generateWarp")
else:
print("Warp initialization successful")
except Exception as e:
print(f"Exception during test buildMesh: {e}")
# Process all cases
results_info = []
for particle_file, orig_mesh_file in cases:
target_particles = sw.ParticleSystem([particle_file]).ShapeAsPointSet(0)
case_id = os.path.basename(particle_file).replace(".particles", "")
print(f"\nProcessing: {case_id}")
print(f"Target particle count: {target_particles.shape}")
# Particle count consistency check
if ref_particles.shape != target_particles.shape:
print(f"Particle count mismatch! Reference: {ref_particles.shape}, Target: {target_particles.shape}")
results_info.append((particle_file, None, None, None, None, "Particle count mismatch"))
continue
# Build warped mesh
warped_mesh = warper.buildMesh(target_particles)
print(f"Warped mesh - Points: {warped_mesh.numPoints()}, Faces: {warped_mesh.numFaces()}")
if warped_mesh.numPoints() == 0 or warped_mesh.numFaces() == 0:
print("Warped mesh is empty")
results_info.append((particle_file, 0, 0, None, None, "Warp failed"))
continue
# Save warped mesh
output_path = os.path.join(output_dir, f"{case_id}_warped.vtk")
warped_mesh.write(output_path)
print(f"Saved: {output_path}")
# Load original mesh and calculate errors
orig_mesh = sw.Mesh(orig_mesh_file)
if orig_mesh.numPoints() == 0:
print(f"Original mesh is empty: {orig_mesh_file}")
results_info.append((particle_file, warped_mesh.numPoints(), warped_mesh.numFaces(), None, None, "Original mesh empty"))
continue
# Calculate point-to-cell distances
dists_and_indexes = warped_mesh.distance(target=orig_mesh, method=sw.Mesh.DistanceMethod.PointToCell)
distances = np.array(dists_and_indexes[0])
avg_err = np.mean(distances)
max_err = np.max(distances)
print(f"Reconstruction error - Avg: {avg_err:.4f} mm, Max: {max_err:.4f} mm")
results_info.append((particle_file, warped_mesh.numPoints(), warped_mesh.numFaces(), avg_err, max_err, "Success"))
# Topology consistency check
print("\nTopology & Error Summary")
if not results_info:
print("No valid warped meshes generated")
else:
base_verts, base_faces = None, None
consistent_topology = True
for item in results_info:
pf, verts, faces, avg_err, max_err, status = item
case_id = os.path.basename(pf)
print(f"{case_id}: Points={verts}, Faces={faces}, AvgErr={avg_err:.4f}, MaxErr={max_err:.4f}, Status={status}")
if status == "Success":
if base_verts is None:
base_verts, base_faces = verts, faces
elif verts != base_verts or faces != base_faces:
consistent_topology = False
if consistent_topology:
print("All successful warps have consistent topology")
else:
print("Inconsistent topology detected in warped meshes")
# Error statistics
failures = [item for item in results_info if item[5] != "Success"]
if failures:
print("\nFailure summary:")
for item in failures:
pf, _, _, _, _, status = item
case_id = os.path.basename(pf)
print(f"{case_id}: {status}")
else:
print("All cases warped successfully")
Mesh Grooming
Fill Holes: enabled
Smooth: disabled
Remesh: enabled
Percentage: enabled
Number of Vertices: 100.0%
Adaptivity: 1.00
Alignment
Reflect: disabled
Alignment: enabled
Method: Center
Reference: auto
Random Subset Size: auto
Other
Skip Grooming: disabled
Optimization Parameters
Number of Particles: 6000
Initial Relative Weighting: 0.05
Relative Weighting: 1
Starting Regularization: 1000
Ending Regularization: 10
Iterations per Split: 1000
Optimization Iterations: 5000
Geodesic Distance: disabled
Geodesic Remesh Percent: 100 (inactive)
Normals: disabled
Normals Strength: 10 (inactive)
Geodesic Distance from Landmarks: disabled
Landmark Distance Weight: 1 (inactive)
Procrustes: disabled
Procrustes Scaling: disabled
Procrustes Rotation/Translation: enabled (grayed out / fixed)
Procrustes Interval: 10 (inactive)
Multiscale Mode: disabled
Multiscale Start: 32 (inactive)
Use Initial Landmarks: disabled
Narrow Band: 4
Best regards,
Lucy